MetaKG: Metabolite-Centric Knowledge Graph Unveils Metabolite Insights Through Multimodal Representation Learning
🧬 Bridging the gap between metabolomics databases through the power of knowledge graphs
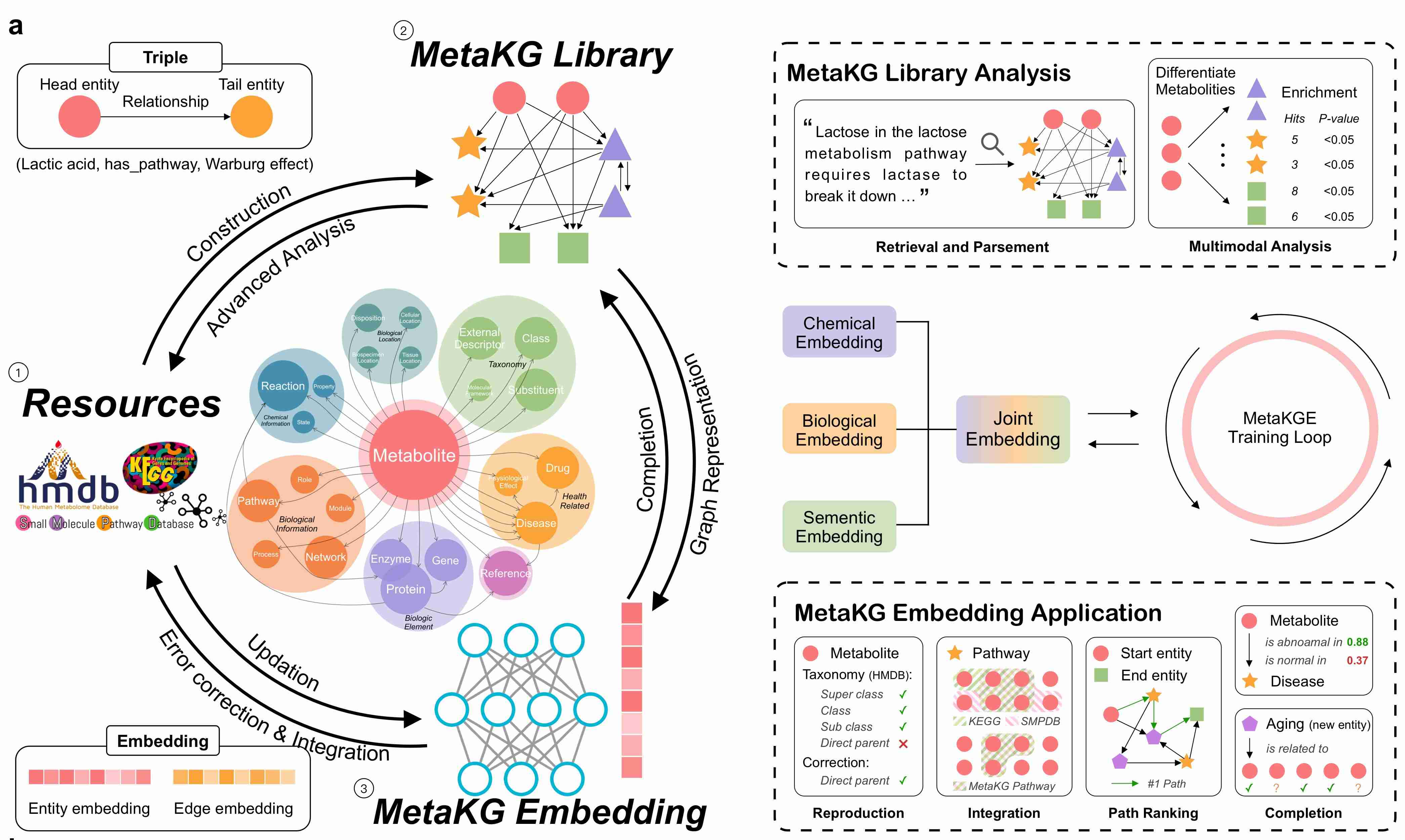
MetaKG is a comprehensive knowledge graph framework that integrates metabolomics data from multiple databases (HMDB, SMPDB, KEGG) into a unified knowledge representation. It provides tools for data integration, analysis, and machine learning on metabolomics data.
Demonstration Website: http://www.metakg.xyz
- 🎯 Unified Access: Query across HMDB, SMPDB, and KEGG with a single interface
- 🔍 Smart Analysis: From basic statistics to advanced machine learning
- 🚀 Production Ready: Battle-tested with large-scale metabolomics data
- 🛠️ Extensible: Easy to add new databases and features
- Unified schema across HMDB, SMPDB, and KEGG
- Automated data extraction and preprocessing
- Cross-database entity alignment
- Multi-format support (XML, JSON, CSV)
- Statistical analysis and visualization
- Network analysis and path search
- Enrichment analysis
- Sankey diagrams for pathways
- KG embedding models (TransE/D/H/R, RotatE, ComplEx, ConvE, etc.)
- Model evaluation suite
- Hyperparameter optimization
- Custom loss functions
- Simple data extraction interface
- Flexible queries
- Batch processing
- Extensible design
- 🎮 CUDA-compatible GPU (recommended)
- 💾 RAM: 16GB minimum, 32GB recommended
- 💽 Storage: 50GB free space
# Core dependencies
python==3.8
pandas==2.0.3
pykeen==1.10.2
bioservices==1.11.2
networkx==3.1
torch==1.9.0
# Optional dependencies
matplotlib==3.7.1
seaborn==0.12.2
scikit-learn==1.2.2
tqdm==4.65.0- Clone the repository:
git clone https://github.com/YuxingLu613/MetaKG.git
cd MetaKG- Download data:
# Download from Google Drive
https://drive.google.com/drive/folders/1TiUtBCG4e2rJ7WIBf_NZ6En8VLbw2aoY
# Place files in the following structure:
data/
├── resource/
│ ├── HMDB/
│ │ └── hmdb_metabolites.xml
│ ├── SMPDB/
│ │ ├── smpdb_metabolites/
│ │ └── smpdb_proteins/
│ └── KEGG/
└── extract_data/- Run example:
python quick_start.pyfrom src.metakg_construction import extract_hmdb_data, extract_smpdb_metabolite_data
# Extract HMDB data
hmdb_entities, hmdb_triples = extract_hmdb_data(
file_path="data/resource/HMDB/hmdb_metabolites.xml"
)
# Extract SMPDB data
metabolite_entities, metabolite_triples = extract_smpdb_metabolite_data(
metabolite_files_dir="data/resource/SMPDB/smpdb_metabolites"
)
# Save processed data
save_entities(hmdb_entities, "data/extract_data/HMDB/hmdb_entities.csv")
save_triples(hmdb_triples, "data/extract_data/HMDB/hmdb_triples.csv")from src.metakg_analysis import summary, search, visualize_graph
# Generate comprehensive statistics
summary.summary(
metakg_library_triples,
show_bar_graph=True,
save_result=True,
topk=20
)
# Search for disease-related pathways
results = search.search_backward(
triples=triples,
nodes=["disease:Nonalcoholic fatty liver disease"],
relations=["has_disease"],
show_only=100
)
# Visualize subgraph
visualize_graph(
triples=results,
save_path="outputs/disease_subgraph.html"
)from src.metakg_machine_learning import kge_training_pipeline, data_partition
# Prepare data
train_path, valid_path, test_path = data_partition.split_data(
triples=metakg_library_triples,
info_path="data/kge_training/info.txt"
)
# Train model
results = kge_training_pipeline.trainging_pipeline(
model_name="RotatE",
loss="marginranking",
embedding_dim=128,
lr=1.0e-3,
num_epochs=2000,
batch_size=16384
)from src.metakg_inference import predict
# Predict diseases related to a metabolite
predictions = predict(
model_name="RotatE",
head="hmdb_id:HMDB0000001",
relation="has_disease",
tail=None,
show_num=3
)
# Predict metabolites related to a disease
predictions = predict(
model_name="RotatE",
head=None,
relation="has_disease",
tail="disease:Diabetes",
show_num=3
)# Example of custom training configuration
config = {
"model": "RotatE",
"loss": "marginranking",
"embedding_dim": 128,
"batch_size": 16384,
"learning_rate": 1e-3,
"num_epochs": 2000,
"negative_sample_count": 50,
"regularization_weight": 1e-5
}
results = kge_training_pipeline.training_pipeline(**config)# Generate Sankey diagram
from case_study.sankey_plot import create_sankey_plot
create_sankey_plot(
triples=metakg_library_triples,
select_relations=['has_pathway', 'has_disease'],
hmdb_list=hmdb_list,
num_relations_to_select=10
)
# Generate enrichment plot
from case_study.MESA_enrichment_plot import create_mesa_plot
create_mesa_plot(
triples=metakg_library_triples,
hmdb_abundance=hmdb_abundance,
select_relations=select_relations,
num_relations_to_select=10
)metakg/
├── src/ # Source code
│ ├── metakg_construction/ # Data extraction & integration
│ │ ├── HMDB/ # HMDB data processing
│ │ ├── SMPDB/ # SMPDB data processing
│ │ └── KEGG/ # KEGG data processing
│ ├── metakg_analysis/ # Analysis tools
│ │ ├── statistics/ # Statistical analysis
│ │ ├── search/ # Path search
│ │ └── visualize/ # Visualization tools
│ ├── metakg_machine_learning/ # ML models
│ │ ├── data_partition/ # Data splitting
│ │ └── kge_training/ # Model training
│ └── metakg_inference/ # Prediction tools
├── data/ # Data storage
│ ├── resource/ # Raw data
│ └── extract_data/ # Processed data
├── case_study/ # Example notebooks
│ ├── sankey_plot.ipynb # Sankey diagram examples
│ └── MESA_enrichment_plot.ipynb # Enrichment analysis
└── checkpoints/ # Model checkpoints
- Smart batch processing for large graphs
- GPU memory management strategies
- Advanced gradient checkpointing
- Efficient negative sampling
- Intelligent early stopping
- Distributed training across GPUs
-
CUDA Out of Memory
- Reduce your batch size
- Enable gradient checkpointing
- Offload to CPU when needed
-
Slow Training
- Check GPU utilization
- Optimize data loading
- Enable mixed precision training
-
Poor Convergence
- Adjust learning rate
- Modify negative sampling
- Try different model architectures
Got ideas? We'd love your help! Here's how:
- Fork the repository
- Create your feature branch
- Make your changes
- Submit a pull request
If you use MetaKG in your research, please cite:
@article{metakg2024,
title={TO BE ADDED},
author={Lu, Yuxing},
journal={TO BE DECIDED},
year={2024},
volume={1},
number={1},
pages={1-10}
}This project is licensed under the MIT License - see the LICENSE file for details.
- Author: Yuxing Lu
- Email: yxlu0613@gmail.com
- GitHub Issues: For bug reports and feature requests
- Discussion Forum: For general questions and community support
- HMDB database
- SMPDB database
- KEGG database
- PyKEEN development team