-
Notifications
You must be signed in to change notification settings - Fork 3
Step 3 CiliaQ Detecting and quantifying cilia from the segmented image
Here we provide an overview about the third step in the CiliaQ workflow: using the "CiliaQ" plugin to quantify the segmented image and retrieve results for all cilia in the image.
CiliaQ reconstructs the ciliary objects based on the image prepared by CiliaQ Preparator and CiliaQ Editor, quantifies them, and visualizes the results.
Tutorial written by Muskan Gupta and Jan N. Hansen in December 2021.
Do you want to take this tutorial as a video?
No: Just scroll down to read the written tutorial.
Yes: Watch it on YouTube
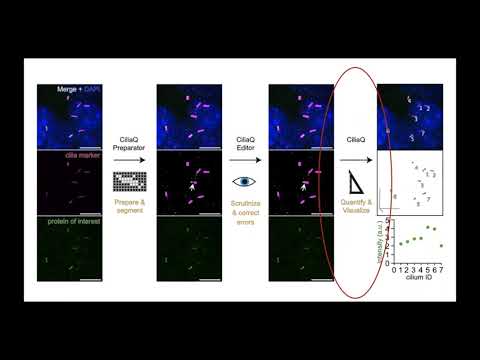
Use CiliaQ to analyze the “_CQP.tif” image from CiliaQ Preparator or, if the CQP image has been edited, the “_CQP_ed.tif” image from CiliaQ Editor with CiliaQ.
To launch CiliaQ, go to Plugins > CiliaQ > CiliaQ... in your FIJI / ImageJ.
A first dialog will pop up for adjusting more general settings:

In the following we explain what each setting in dialog means:
Process: There are 3 options available.
A) Multiple Images: If you select this option, the multiple images dialog will pop up as soon as you have finished setting all analysis parameters in the dialogs and will allow you to create a list of files to be processed.

All you need to do is adding your files to the list which you can do in two ways:
- Select files individually.
- Select files by pattern - select a folder and list those files from the folder that match a certain file name pattern.
By either method you will generate a list of files.

By start processing the analysis will start.
B) Active image in FIJI: This option will provide you the opportunity to process only the currently opened front-most image in FIJI.
C) All images open in FIJI: Here all the images that are open in FIJI will be processed, one after the other.
Note: Generally, when subjecting multiple images to CiliaQ Preparator, the plugin will process the images one after the other. This “batch-processing” is particularly helpful for large data sets, as it allows you to run, e.g., 100 images just by one click and then, as no further user interaction is required, you can spend the time while the images are processed with something else.
Calibration settings: If the image does not contain any calibration information, check the check box “manually calibrate image” and enter the calibration information for all images to be analysed.

Note: You can check the calibration information for an opened active image in FIJI via Image > Properties. If the image is calibrated you see a unit like “micron” written (if it is not calibrated, there will be "pixel" written).
Properties dialog for a calibrated image (left) and an uncalibrated image (right).


Alternatively, you can see whether the image is calibrated in the status bar of an opened image on the top of the image window:

Detection settings: There are 2 options available. A) Manually enter preferences: If you select this option, you can enter preferences manually in a second dialog that pops up after confirming this dialog. B) Load preferences from existing CiliaQ Preparator metadata file:

Both ways are explained below:
A) Manually enter preferences: If you select this option, you can enter preferences manually in a second dialog that pops up after confirming this dialog.

- Intensity quantifications: Set here whether you want to quantify any intensity values or cilia coverage in additional channels of the multi-channel image:

- Basal stain: If a channel in your image provides a basal stain, check and specify the channel number:

Note: The intensity information from basal stain is used to orient the output intensity profiles from base to tip.
- Channel specification: Specify here what can be found in which channel of the image.

The setting shown in this image means that
The first channel (segmented channel): contains cilia labeling and serves for reconstructing the cilia.
The second channel: quantified for intensities and referred to in the output files as intensity “A”
The third channel: quantified for intensities and referred to in the output files as intensity “B”
- Note: Here, the channel position in the “_CQP.tif” or “CQP_ed.tif” image need to be specified – do not specify the position here that corresponds to the channel position in the raw microscopy file! If you are not sure, which channel is which channel nr in the CQP.tif file, have a look at the CQP.txt file and you will find a list which channel from the raw image file has ended up in which channel in the CQP.tif file.

-
Object detection / connection: The increase range function
 determines which mode is used to connect adjacent voxels to form a cilium object.
determines which mode is used to connect adjacent voxels to form a cilium object.
- If the box is checked, CiliaQ will connect a voxel in the image to all direct (blue voxels in image below) and diagonally adjacent voxels (orange voxels in image below) to form a cilium object.
- If the box is not checked, diagonally adjacent voxels (orange in image below) will not be connected but only directly adjacent voxels (blue).

- The “increase range” function is helpful when the image features a bad signal-to-noise-ratio or when cilia are incompletely labeled and contain small gaps in labeling.
- However, the “increase range” function also features the disadvantage that noise voxels can be added to cilium objects and can cause erroneous length detection.
- Example:
Segmented channel (white: positive, “ciliary” voxels, black: background voxels):

Detected cilium object when “increase range” option is selected:

Detected cilium object when “increase range” option is not selected:

Image sources: Hansen, et al. CiliaQ: a simple, open-source software for automated quantification
of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E_ 44, 18 (2021).
doi: 10.1140/epje/s10189-021-00031-y. Licensed under CC BY 4.0.
-
Noise filtering: Estimate the minimum size of a cilium: All cilium objects containing less voxels than the minimum specified here will be considered as noise and removed from the image:

-
Removing incomplete objects at the image borders: CiliaQ allows to exclude objects that do not lie completely within the imaged field of view. Objects contacting the image borders in x, y, and z are automatically removed if the following option is set as follows:

- Note: In case you are analysing a 2D image, change the exclude option to “cilia touching x or y borders”, because otherwise no cilia will be analyzed as any object will be interpreted as touching the z border:

- Segmented intensity channels: CiliaQ allows to analyze channels as “intensity A” or “intensity B” channels that are segmented like the reconstruction channel. This allows to determine the overlap of the ciliary 3D object with objects in these segmented intensity channels. To this end, CiliaQ also offers to perform noise removal and specify whether obejcts detected in additional channels than the reconstruction channel shall be detected with or without the “increase range” function:

-
Note: Leave the ”minimum size of intensity regions (for A and B)” at 1 if you do not provide segmented intensity channels.
-
Skeleton-based results: Skeletonization is used to determine the ciliary center line. The skeleton determines the length of the cilium and intensity profiles along the central track (“skeleton”) of the cilium. To avoid that the skeleton gets side branches or additional kinks (falsifying the results), a Gauss filter is applied before skeletonization to smoothen the surface of the ciliary object. The Gauss filter should be used when length estimations do not properly work (e.g. skeletons show side kinks or >1 branches).

- Note: Usually, a Gauss filter is applied only in XY and at sigma values of 1 or 2. Make sure you use the identical sigma value for all data from the same dataset. Total length values might slightly vary among results from different sigma (see also Fig. 1 of the original CiliaQ publication).

Source: Hansen, et al. CiliaQ: a simple, open-source software for automated quantification
of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E_ 44, 18 (2021).
doi: 10.1140/epje/s10189-021-00031-y. Licensed under CC BY 4.0.
-
Tangent and curvature calculation: CiliaQ calculates and outputs the tangent vector and the local curvature for each ciliary point.
- How are tangents determined and curvatures calculated?
- For each ciliary skeleton point: the tangent is formed as the vector from the point at a specific arc length distance upstream on the cilium to the point at a specific arc length distance downstream on the cilium. The tangent vector is then normalized to a length of unity. This specific distance of the point for which the tangent vector is calculated to the points used to calculate the tangent vector is set here.

- Tangent vectors up- and downstream of the point of interest are used to determine the curvature. The distance between the two points used is defined by the parameter setting, as well.
- Note: The parameters tangent vector and curvature are relevant only for specific applications in the physics community – so if you apply CiliaQ for standard applications, leave the setting at the default value.
- How are tangents determined and curvatures calculated?
B) Load preferences from existing CiliaQ file:
- If you select this option, you can select a CiliaQ output file later (ending “..._CQ.txt”) to import settings: a second dialog will pop up after confirming this dialog to select the metadata file.


- After loading, the Log window will pop up and display the loaded preferences. It allows you to assure loading worked correctly.

- Note: If you open a file from a CiliaQ analysis with CiliaQ versions before v0.1.4, you are requested by another dialog to add the missing parameter for tangent and curvature calculation.
Output settings allow to retrieve more output files or to add a data suffix in the output file names.

-
Save date in output file names: It is helpful in avoiding that a CiliaQ analyses overwrites already existing CiliaQ results files from a different CiliaQ analysis on the same image. Example: If you want to compare the CiliaQ results obtained by different settings.
-
Save result images for each individual cilium: It creates a folder containing stack images for each detected cilium. Example: Folder name = name of the analysed image name + suffix “_CQ”.
- It is necessary to check this option if you want to look at individual cilia in time-lapse images, as for time-lapse images the common output files will just give averaged results for all cilia. However, if you select this option, you get output files for individual cilia in time lapse images as well. If you did not use time-lapse images, the common output files provide all information for each cilium separately and there is no need to activate this checkbox for standard applications.
-
Save 3D visualizations for each individual cilium: It saves 3D visualizations for each individual cilium in the same folder as result images for each individual cilium. Example: Folder name = name of the analyzed image name + suffix “_CQ”
- Note: 3D visualizations can slow down the analysis pipeline because their generation is time-consuming. If you analyze many cilia and create a 3D projection for each cilium this will take very long.
-
Save 3D visualizations for whole image:
- If 3D visualization of the noise-filtered reconstruction channel is done, then the file will be saved as filename + suffix "_CQ_RP_3D.tif”
- If 3D visualization of the detected skeletons is done, then the file will be saved as filename + suffix "_CQ_SKL_3D.tif”
-
Output number format: Decide between the number format of US (0.00) or Germany (0,00). Note: If you use excel for further data analysis, select this depending on how your excel shows digits.
All output files from CiliaQ for a given input image are saved in the directory of the input image and contain the name of the analysed input image plus the suffix “_CQ” and, eventually, extra-suffixes depending on the type of output file.
CiliaQ always outputs the following files:
- Filename + suffix "_CQ.txt": A tab-delimited text file containing the analysis settings and results
- Filename + suffix "_CQ_RP.tif": A copy of the input image after noise filtering the reconstruction (and, if applicable, the intensity region) channel(s). This image also contains labels of the detected cilia objects (labels correspond to the column ID in the output text files. This allows you to easily relate analysis results from the text file to a specific cilium in the image.
- Filename + suffix "_CQ_SKL.tif": An Image (stack) showing the detected skeletons, labelled with the analysis IDs of the respective cilium objects.
- Filename + suffix "_CQs.txt": A tab-delimited text file containing only the results lines from the file with suffix _CQ.txt, without table caption. Retrieve the caption for that table from the table in the file with suffix _CQ.txt.
To learn more about the CiliaQ Outputs, visit the next tutorial.
Questions? Suggestions? Let us know at the CiliaQ Discussions forum or via email: jan.hansen (at) uni-bonn.de
Copyright (C) 2017-2024: Jan N. Hansen.
CiliaQ is part of the following publication: Jan N. Hansen, Sebastian Rassmann, Birthe Stueven, Nathalie Jurisch-Yaksi, Dagmar Wachten. CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E 44, 18 (2021). https://doi.org/10.1140/epje/s10189-021-00031-y