-
Notifications
You must be signed in to change notification settings - Fork 1
UsingTheCSD
The Cambridge Structural Database (CSD) contains >1 million crystal structures for organic and organometallic molecules. The CSD is part of the CSD Portfolio which also includes software for:
-
Search, retrieval and analysis of structures - ConQuest
-
Crystal structure visualisation and geometric analysis - Mercury
-
Generation of in-house databases searchable alongside the CSD - PreQuest
-
The CSD Portfolio also incorporates IsoStar, a library of intermolecular interactions, containing data derived from both the CSD and PDB, and Mogul, a molecular geometry library.
-
For more information about the Cambridge Crystallographic Data Centre (CCDC)
see: http://www. ccdc.cam.ac.uk
-
The search program ConQuest allows one to retrieve all molecular fragments specified by drawing chemical diagrams. The chemical environment of the fragments can be specified very precisely using a variety of attributes such as number of hydrogens per atom, cyclicity of bonds, etc. The geometry of these fragments may be saved as a list of geometric parameters defined by the user, and displayed as histograms and scattergrams by the program Mercury.
-
DASH provides a direct link to Mogul, a molecular geometry database which forms part of the CSD Portfolio and is available from the CCDC. Searches for molecular fragments, bond lengths, bond angles and torsion angles can be performed quickly and easily.
-
The ConQuest program is also able to search for intermolecular interactions, and store parameters in the same way as for intramolecular geometry, for examination by the Data Analysis module within Mercury. Examples of such useful information are H-bonds and chloride ion interaction with charged nitrogen. The packing motifs of the retrieved structures can be examined using the visualiser Mercury. These programs are particularly useful for easy exploration of H-bonding motifs.
-
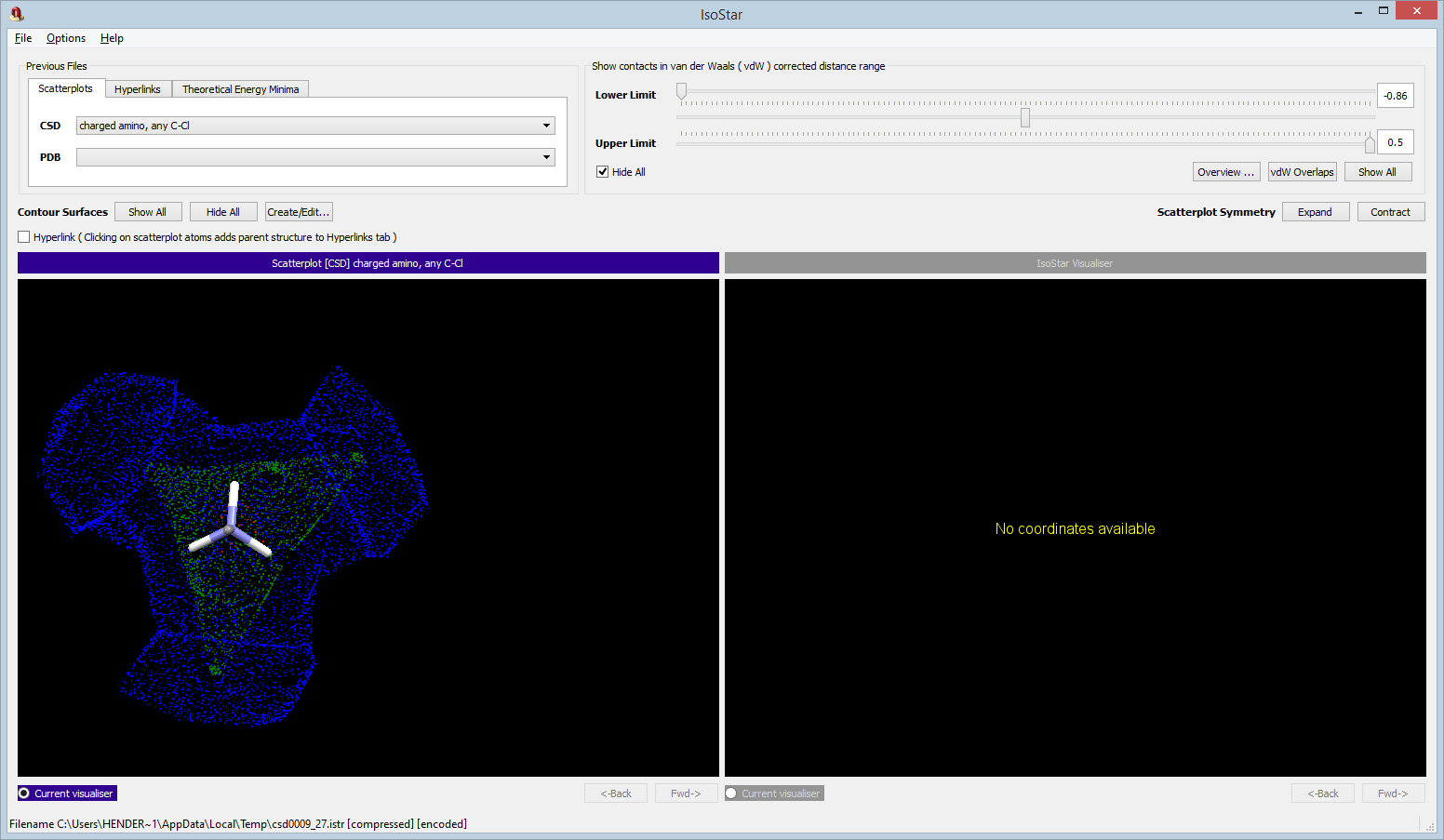
The IsoStar library of intermolecular interactions is also provided within the CSD Portfolio. This is an extensive library of scattergrams of the intermolecular crystal environment of a set of well-known chemical groups. Each group (termed the central group) has a set of pre-processed scattergrams of interacting groups in the CSD, taking account of symmetry to produce an overall picture. These scattergrams can be easily inspected using the visualiser provided.
An IsoStar example is shown here of a central charged amine group, NH3+, approached by a chloride ion, Cl-.

In the IsoStar contour view option, the scatterplot has been contoured to show the preferred positions of the chloride at about 3.0 Å from the nitrogen.